Pathwalker
Pathwalker is a method for building models from cryo-EM density maps described in the following publication:
Pathwalker components
The individual Pathwalker components have wiki pages with usage, help, and FAQs:
Example
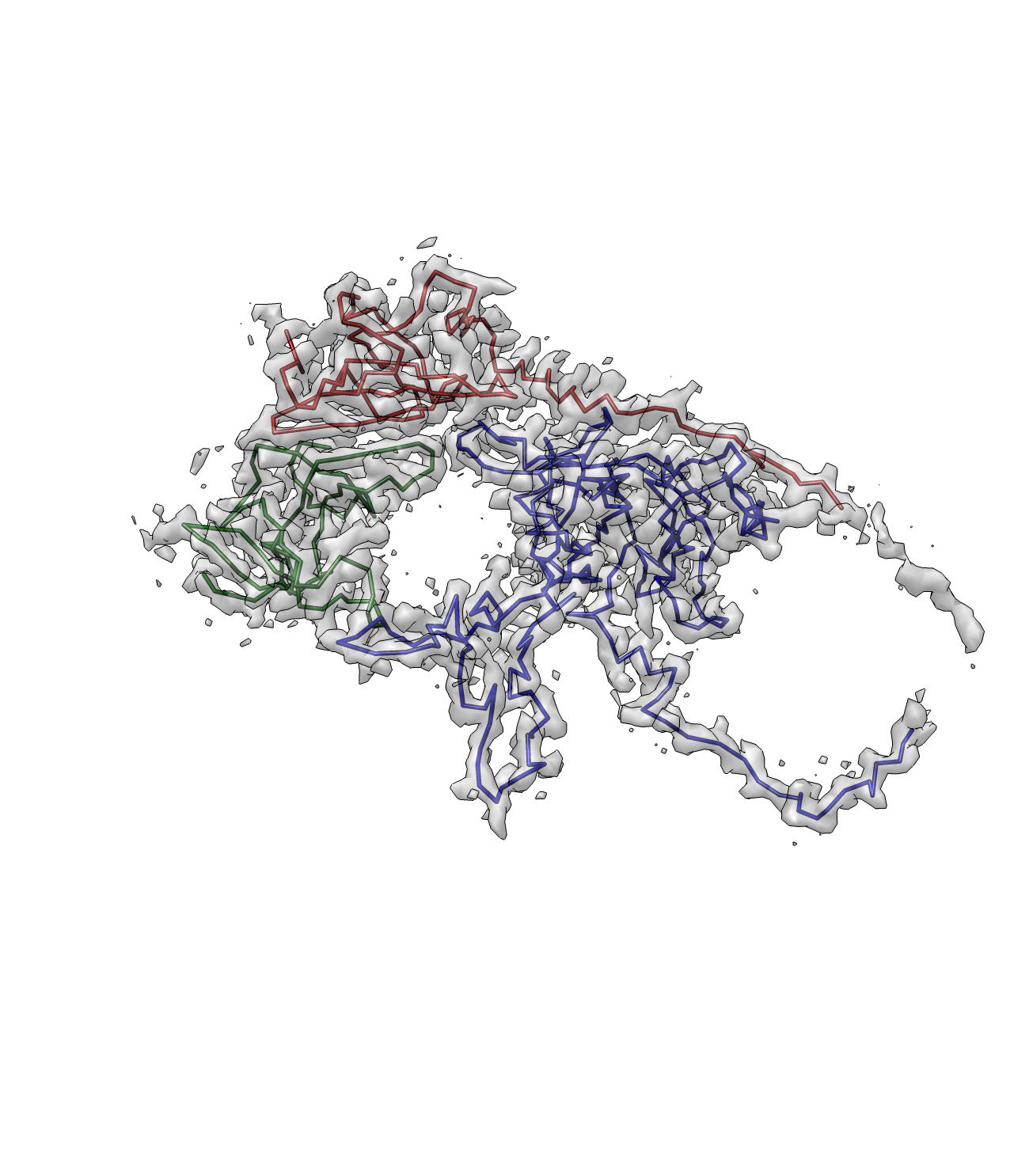
Rotavirus:
First segment out the density of one subunit. Name it "rotav.mrc"
Then threshold out the negative density in the map using: (This is important, or it will cause trouble in the TSP solver) e2proc3d.py rotav.mrc rotav.mrc --process=threshold.belowtozero
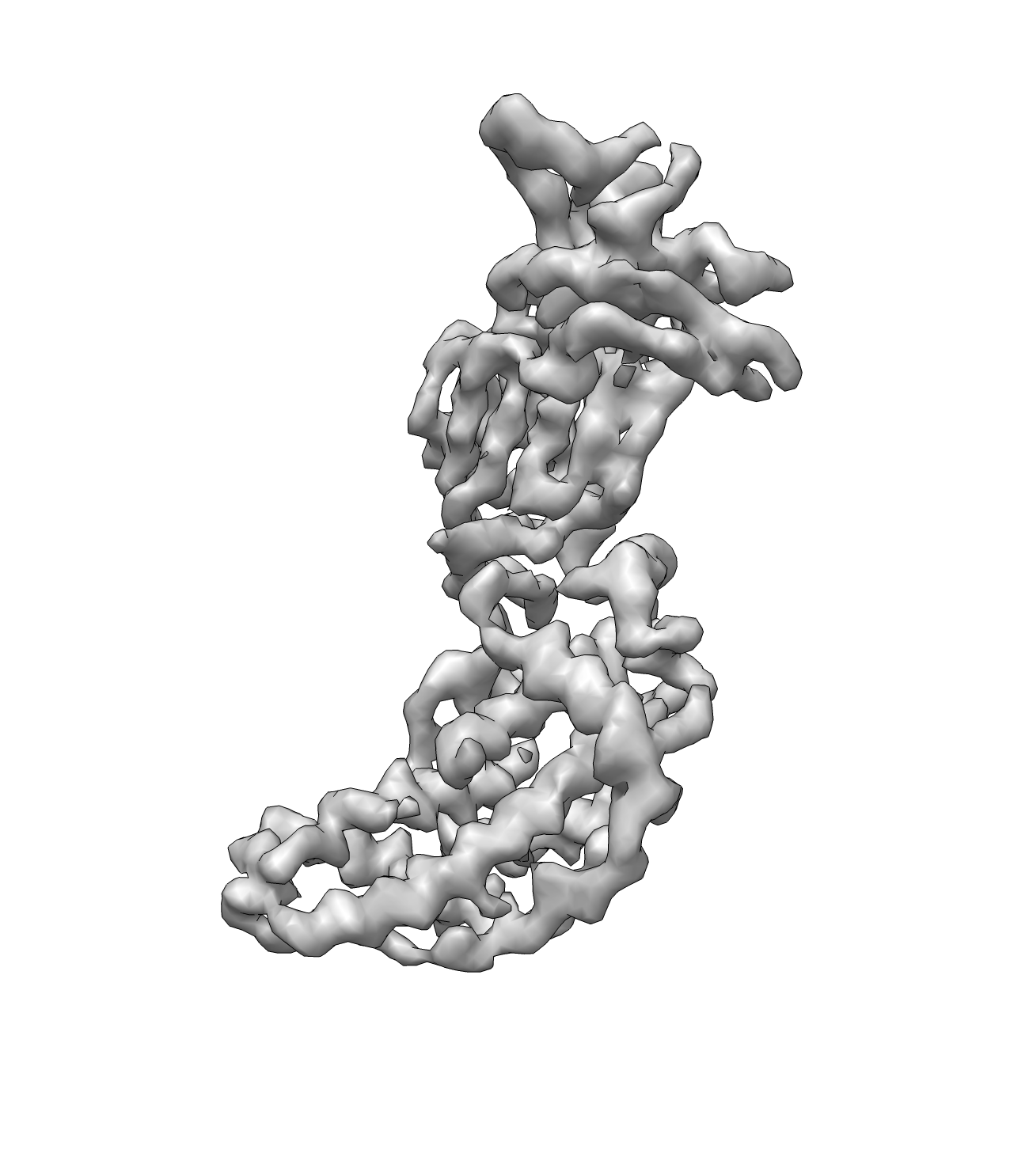
Seed the pseudoatoms using the command:
e2segment3d.py rotav.mrc --pdbout=rotav_pseudoatoms.pdb --process=segment.kmeans:ampweight=1:nseg=398:verbose=1:minsegsep=1:pseudoatom=1:thr=4
Here nseg is required which is the number of residues in the protein. "thr" is the threshold of the isosurface. The program will choose an (hopefully) optimized value for you. Then trace the backbone using the LKH TSP solver.
e2pathwalker.py rotav_pseudoatoms.pdb --mapfile=rotav.mrc --output=rotav_path1.pdb --solver=lkh --overwrite --dmin=2 --dmax=10 --mapthresh=6 --mapweight=100
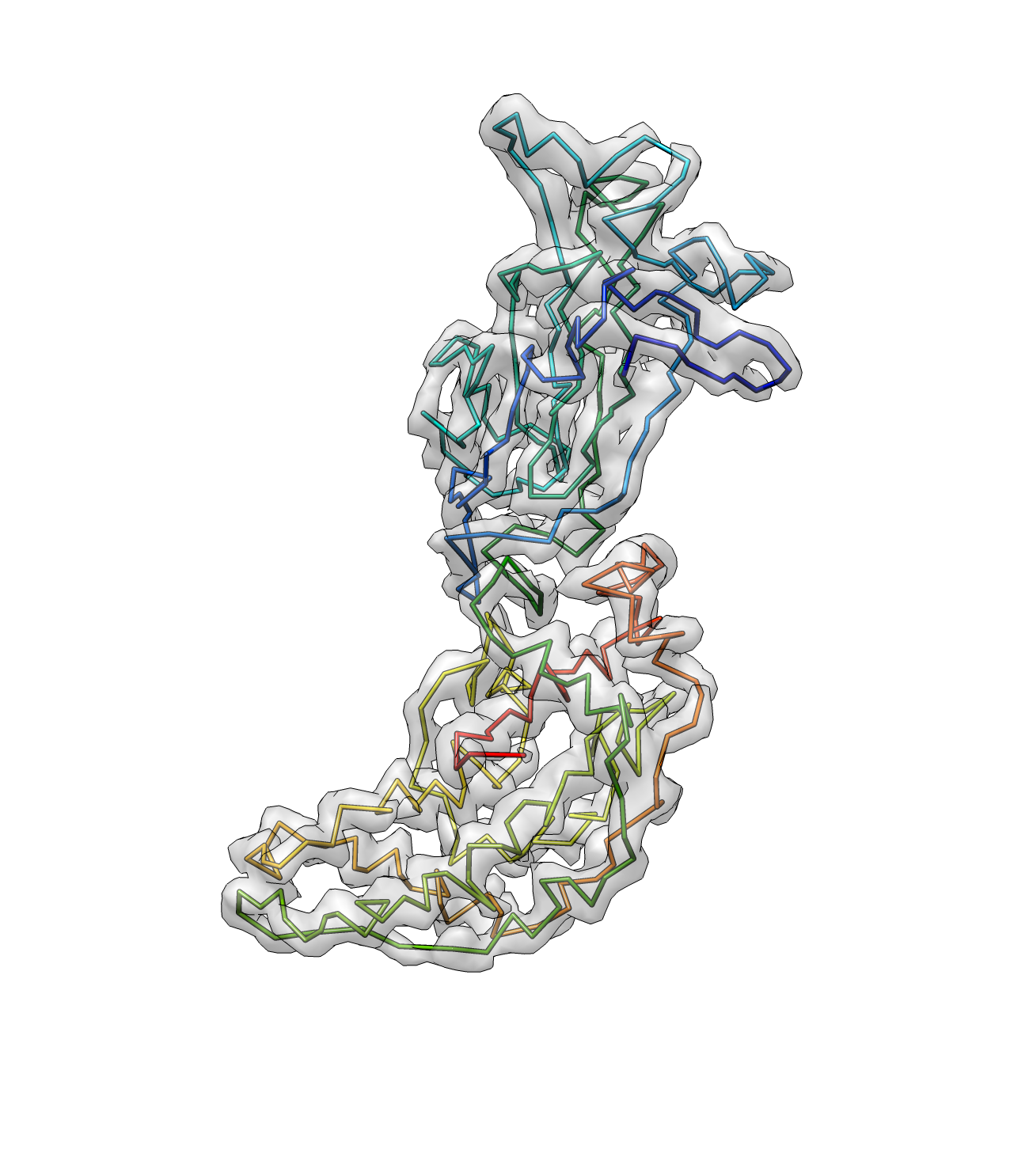
Here dmin and dmax are the minimum and maximum distance between C-alphas. The dmax should be set larger when the map is not well connected. mapthresh is the threshold for the density value in the map, and the bonds with a density higher then the threshold will be weighted more. mapweight is a parameter that weight between the map and protein geometry score. The program will trust the density map more when this value is higher (it then tends to connect two atoms that has strong density connection even the distance is far from the average distance between C-alphas)
The helices can then be found using the command:
e2pwhelixfit.py --mapin=rotav.mrc --pdbin=rotav_path1.pdb --output=rotav_hlx1.pdb --lenthr=12 --denthr=8 --mapwohelix=rotav_rmhlx.mrc --minlen=8
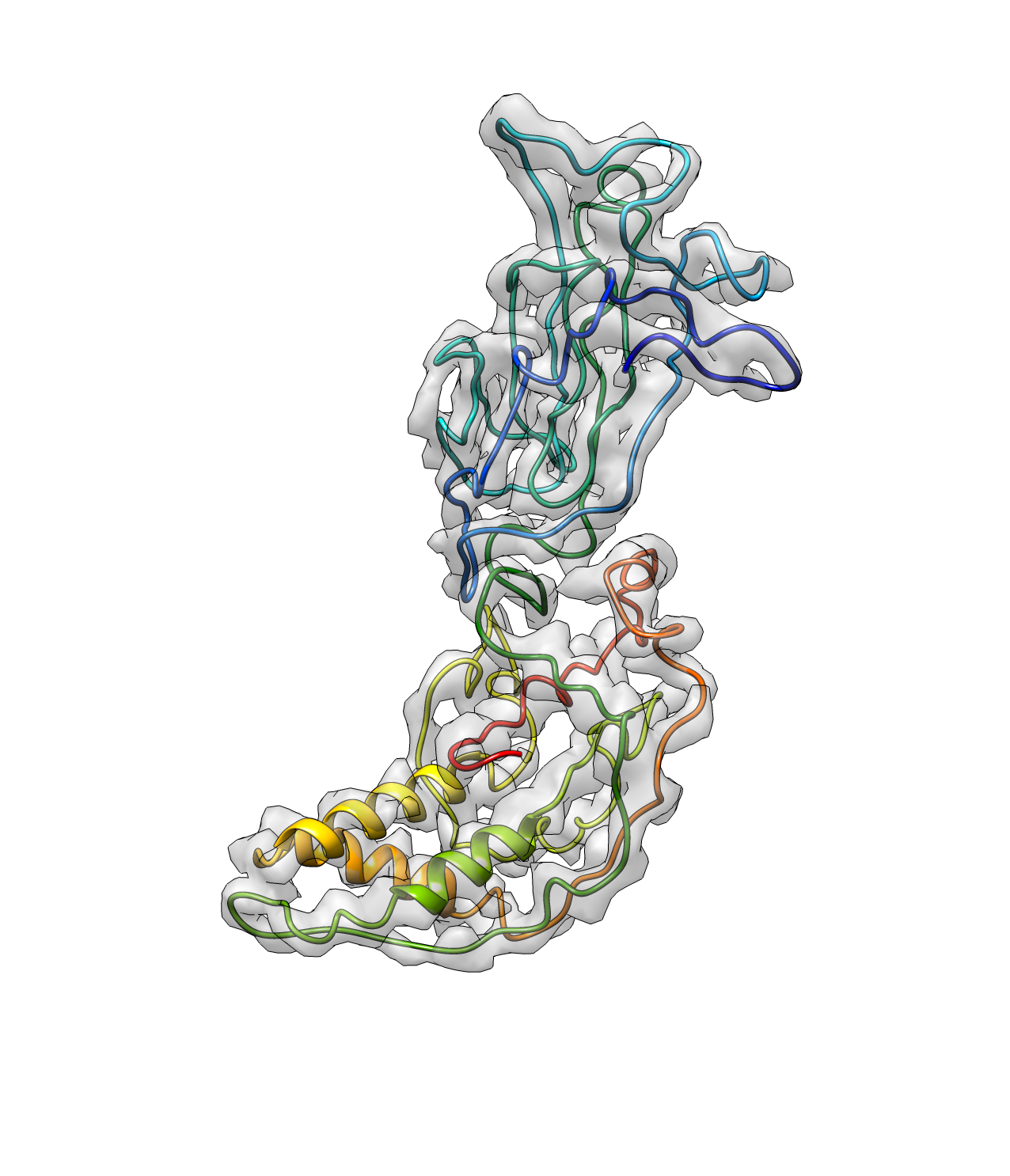
The argument minlen sets the minimum length of the helix and denthr sets the minimun mean density at the position of the atoms in the helix. mapwohelix gives you a map with the density of all the detected helices removed, which will be used for pseudoatom seeding in the next iteration.
Remove the dust voxels(mostly sidechains in the helices) in the map. e2proc3d.py rotav_rmhlx.mrc rotav_hd.mrc --process=mask.dust3d:voxels=200:threshold=6
Do the pseudoatom generation again, using the density map without helices and specify the pdb file that contains the helices.
e2segment3d.py rotav_hd.mrc --pdbout=rotav_psudoatoms2.pdb --process=segment.kmeans:ampweight=1:nseg=398:verbose=1:minsegsep=2:psudoatom=1:thr=6 --helixfile=rotav_hlx1.pdb --edgefile=helix1
Tracing the path. Note that the mapweight is set higher this time because we are focusing on the loops with lower density value and not well connected. e2pathwalker.py rotav_pseudoatoms2.pdb --mapfile=rotav.mrc --output=rotav_path2.pdb --solver=lkh --overwrite --dmin=2 --dmax=10 --mapthresh=6 --mapweight=500 --edgefile=helix1
Then repeat this process:
e2pwhelixfit.py --mapin=rotav.mrc --pdbin=rotav_path2.pdb --output=rotav_hlx2.pdb --lenthr=6 --denthr=8 --mapwohelix=rotav_rmhlx2.mrc --minlen=6
e2proc3d.py rotav_rmhlx2.mrc rotav_hd2.mrc --process=mask.dust3d:voxels=200:threshold=6
e2segment3d.py rotav_hd2.mrc --pdbout=rotav_pseudoatoms3.pdb --process=segment.kmeans:ampweight=1:nseg=398:verbose=1:minsegsep=2:psudoatom=1:thr=6 --helixfile=rotav_hlx2.pdb --edgefile=helix2
e2pathwalker.py rotav_pseudoatoms3.pdb --mapfile=rotav.mrc --output=rotav_path3.pdb --solver=lkh --overwrite --dmin=2 --dmax=10 --mapthresh=6 --mapweight=500 --edgefile=helix2
e2pwhelixfit.py --mapin=rotav.mrc --pdbin=rotav_path3.pdb --output=rotav_hlx3.pdb --lenthr=8 --denthr=8 --mapwohelix=rotav_rmhlx3.mrc --minlen=6
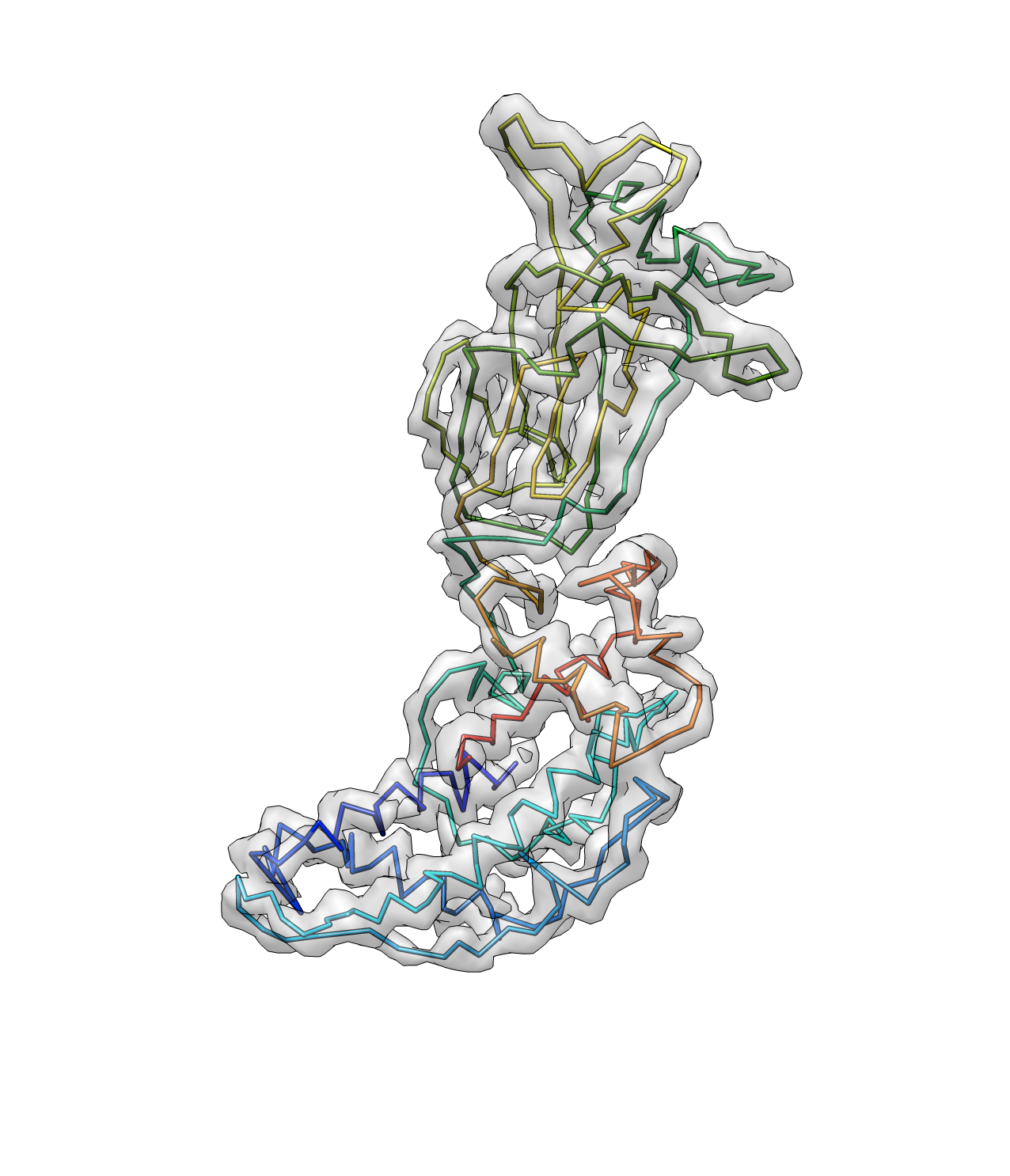
And now we have a better path here. Note that the termini points are now corrected.
These commands will help clean up the crossed bonds, obvious sidechain atoms and some bonds that are too long.
e2pwstructuremodify.py --mode c --mapin=rotav.mrc --pdbin=rotav_path3.pdb --output=rotav_path3_mdf.pdb --thr=1 e2pwstructuremodify.py --mode d --mapin=rotav.mrc --pdbin=rotav_path3_mdf.pdb --output=rotav_path3_mdf2.pdb
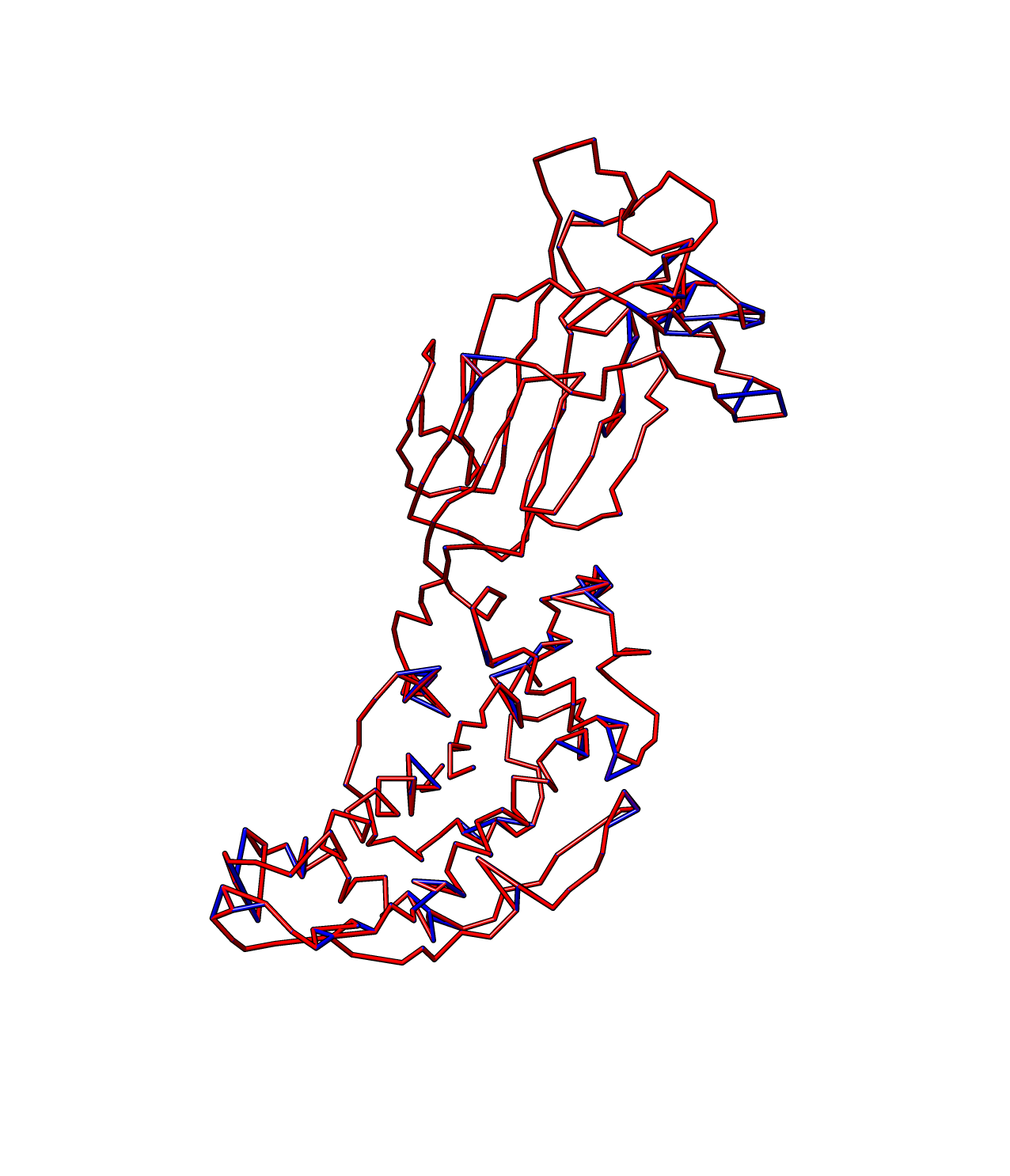
To do some manual modification, like connecting a pair of atoms that's not connected in the model, simply put the the ID of the two atoms at the end of the helixN file like this: ( here is the file helix2) 270 288 264 138
Then run the pathwalker again:
e2pathwalker.py rotav_psudoatoms3.pdb --mapfile=rotav.mrc --output=rotav_path4.pdb --solver=lkh --overwrite --dmin=2 --dmax=10 --average=2 --mapthresh=7.5 --mapweight=500 --edgefile=helix2
The program will rebuild the rest part of the model for you.
To trace multiple subunits, simply add --subunit N in the e2pathwalker.py command and the program will try to find the best point to break the chain.