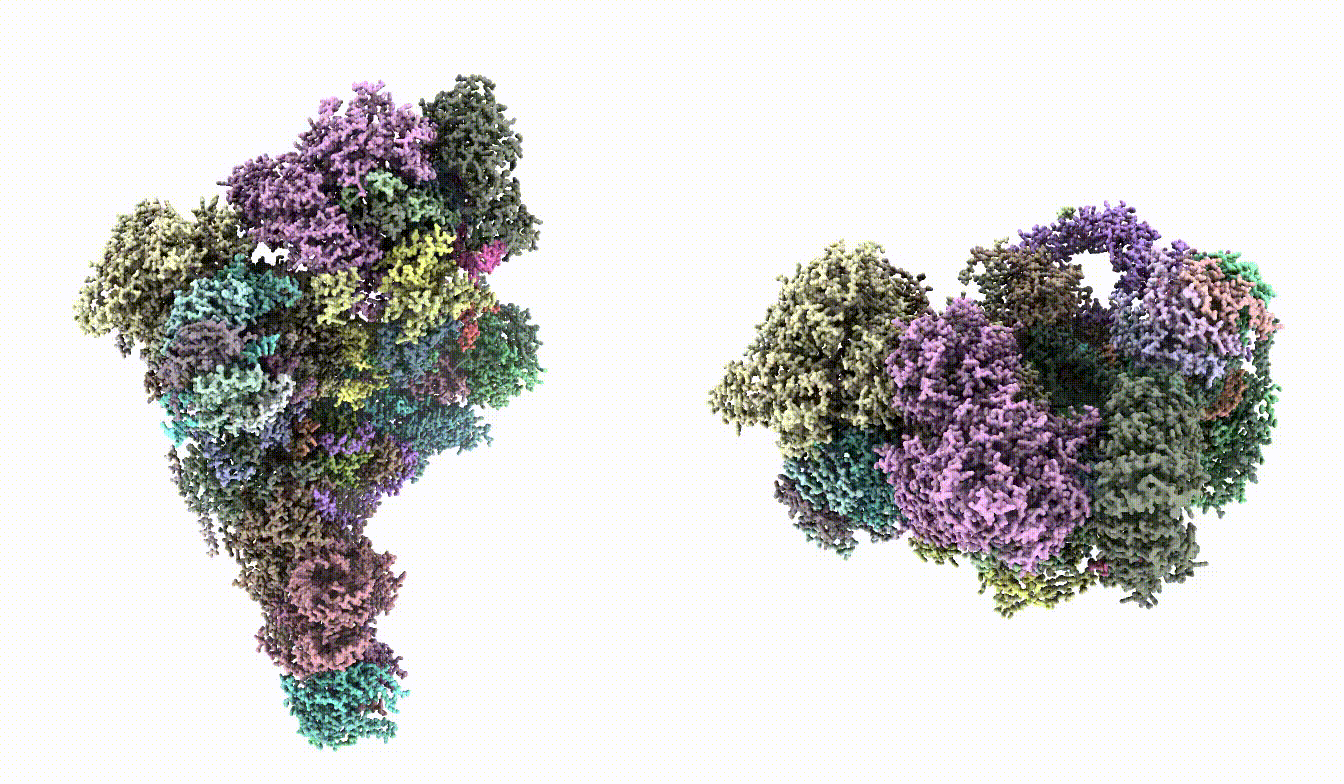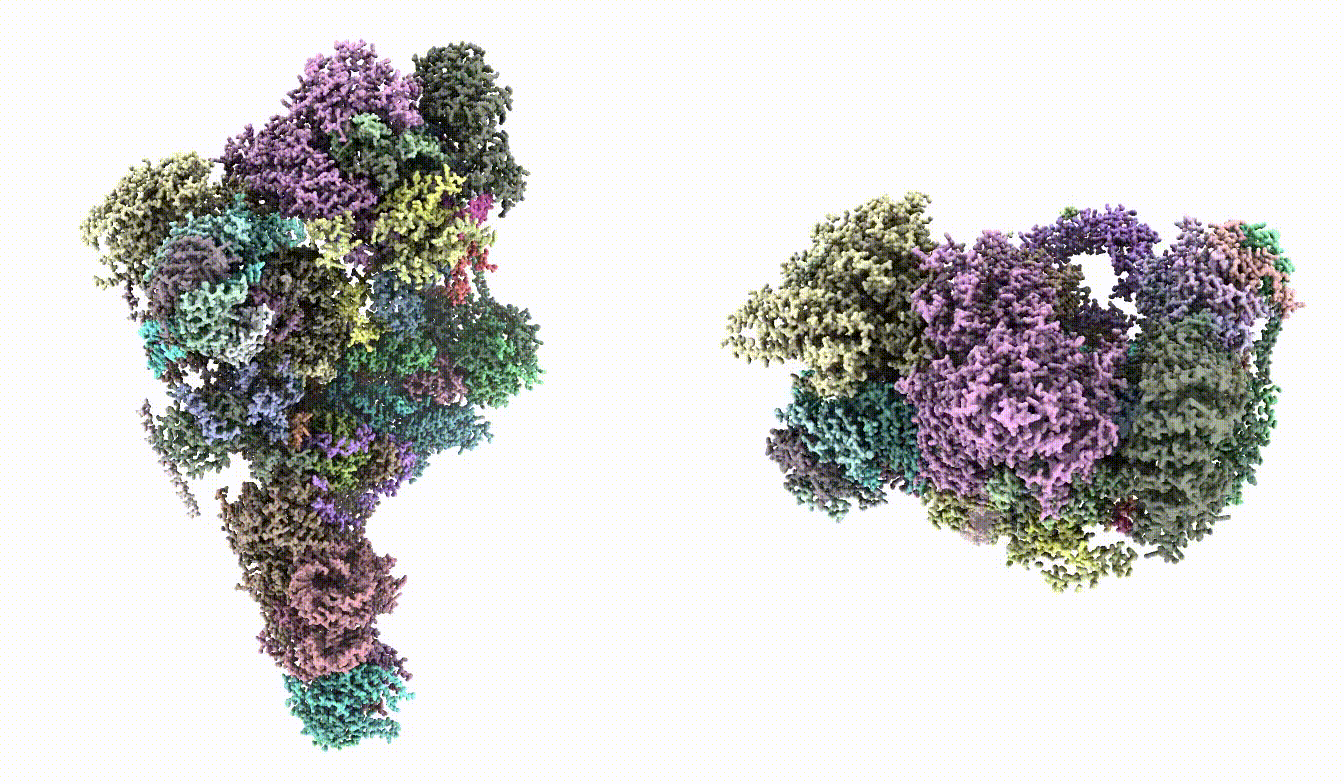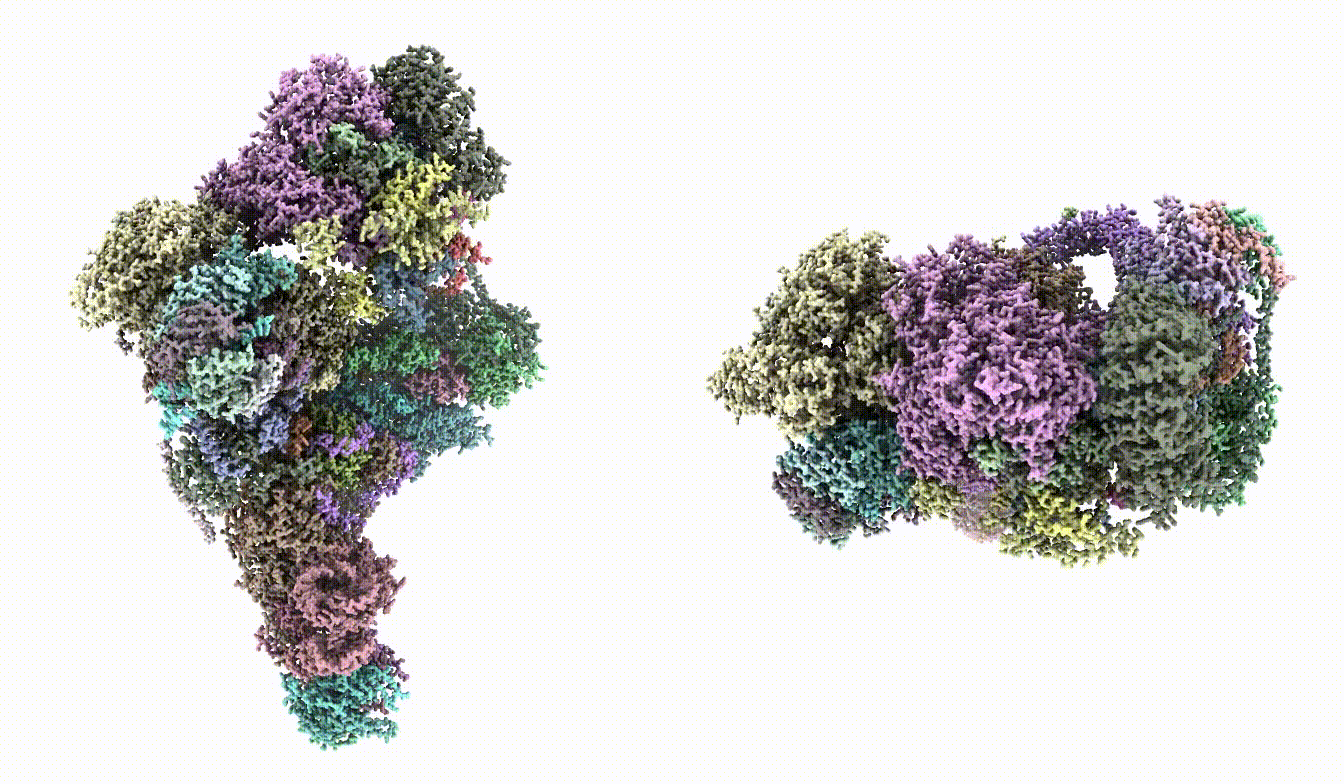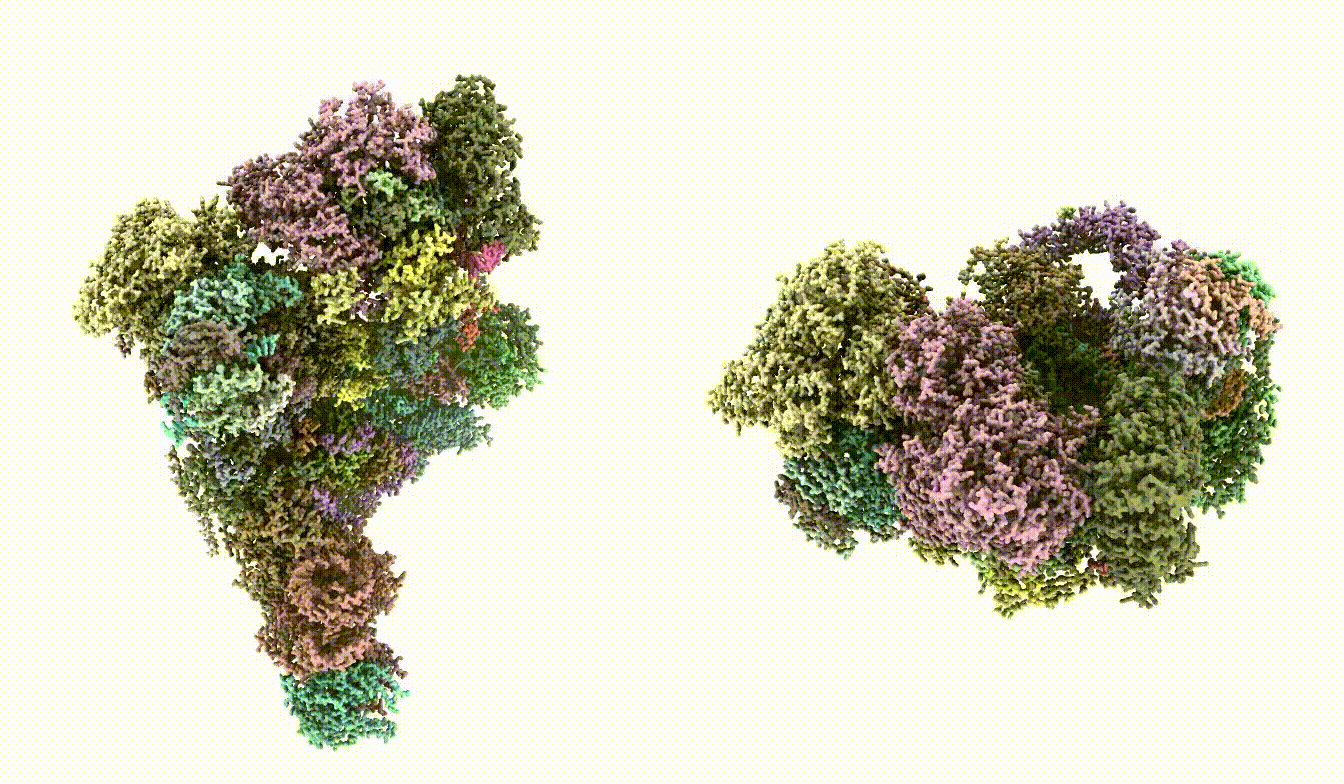|
Size: 1008
Comment:
|
Size: 4111
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 3: | Line 3: |
| Refine atomic models to better fit into a given map, and get better PDB validation score. | Refine atomic models to better fit into a given map, and get better PDB validation score. Gradually updating... == Dataset == In this tutorial, we use TRPV1 as an example. Starting from a relatively old PDB model (PDB: [[https://www.rcsb.org/structure/3J5Q|3J5Q]] ), we will fit it into a newer, higher resolution EMDB structure ( [[https://www.emdataresource.org/EMD-8117|EMD-8117]] ). This is quite similar to the typical modeling work, when we have an existing homolog or predicted model and a newer, better structure. In this case, the starting model was built before the PDB validation system came out, so the validation score is not pretty. {{attachment:pdbval_pdb.png | validation_pdb |width=400}} First, roughly fit the model into the map. This can be done in ChimeraX. They have slightly different conformation, so the fit won't be very precise. It is ok to leave it as is. {{attachment:pdb_fit.png | Fitting model to map |width=400}} |
| Line 7: | Line 20: |
| First we compute some parameters from the input model that will be used in the following refinement. | First, we compute some parameters from the input model that will be used in the following refinement. |
| Line 10: | Line 23: |
| e2gmm_model_compile.py --model model_input.pdb | e2gmm_model_compile.py --model 3j5q.pdb |
| Line 13: | Line 26: |
| Some non-protein structures are not supported at the moment. It is recommended to remove them from input and add them back after refinement. | This should create a `gmm_model_xx` folder, which contains information on the stereochemical constraints of the input model. All output will be in PDB format if the input model here is PDB. If the input is CIF, the output will be in CIF. Otherwise, specifying `--writecif` will force output to be CIF. For your own data, it is worth noting that some non-protein structures are not supported at the moment. It is recommended to remove them from input and add them back after refinement. It is also good to notify me about those so I can add them in the future. |
| Line 18: | Line 33: |
| e2gmm_model_fit.py --path gmm_model_00 --map map_input.hdf --resolution 3 | e2gmm_model_fit.py --path gmm_model_00 --map emd_8117.mrc --resolution 3 --writetxt --rebuild_rotamer |
| Line 21: | Line 36: |
| Here `gmm_model_00` is the folder generated by the `e2gmm_model_comple.py` command. This step can be skipped if there is no input map. | Here `gmm_model_00` is the folder generated by the `e2gmm_model_comple.py` command. This step can be skipped if there is no input map and you only want to refine the stereochemical score of the model. Make sure the model does fit in the map in the first place. In this case, the loss from command line output should start from around -0.4 and gradually decrease to -0.6 or lower throughout iterations. The `--rebuild_rotamer` option will select the rotamer for each residue that fits best into the given map (by map-model FSC). The process can be very slow. It is necessary for this case because there are many sidechain outliers, but can be skipped if the starting model is more reasonable. Take a look at `fit_03.pdb` and the model should fit into the map. {{attachment:fit_map.png | model fit in map |width=400}} |
| Line 30: | Line 54: |
Normally, the default option should produce a good enough model. But in this case, since we started from a quite bad position, it would be necessary to run the command multiple times. For the second run, specify `--niter 0,20` to skip the initial iterations, and `--fixrotamer` to force good rotamers for the bad residues. {{{ e2gmm_model_refine.py --path gmm_model_00 --model gmm_model_00/model_02.pdb --fixrotamer --niter 0,20 }}} There is some randomness in the process, and the command can be run multiple times. In the end, this should give relatively good PDB validation score. {{attachment:pdbval2.png | pdb validation output |width=400}} == Multi-model refinement == After heterogeneity analysis, we can build models for a continuous conformational change using a stack of volumes generated by `e2gmm_eval.py`. {{{ e2gmm_model_multi.py --path gmm_model_00 --modelpdb gmm_model_00/model_02.pdb --modeltxt gmm_model_00/fit_03.txt --maps gmm_00/ptcls_cls_01.hdf --resolution 10 --ndim 2 }}} {{attachment:model_movie.gif | movement movie |width=800}} |
Gaussian mixture model based atomic model refinement (2024)
Refine atomic models to better fit into a given map, and get better PDB validation score. Gradually updating...
Dataset
In this tutorial, we use TRPV1 as an example. Starting from a relatively old PDB model (PDB: 3J5Q ), we will fit it into a newer, higher resolution EMDB structure ( EMD-8117 ). This is quite similar to the typical modeling work, when we have an existing homolog or predicted model and a newer, better structure.
In this case, the starting model was built before the PDB validation system came out, so the validation score is not pretty.
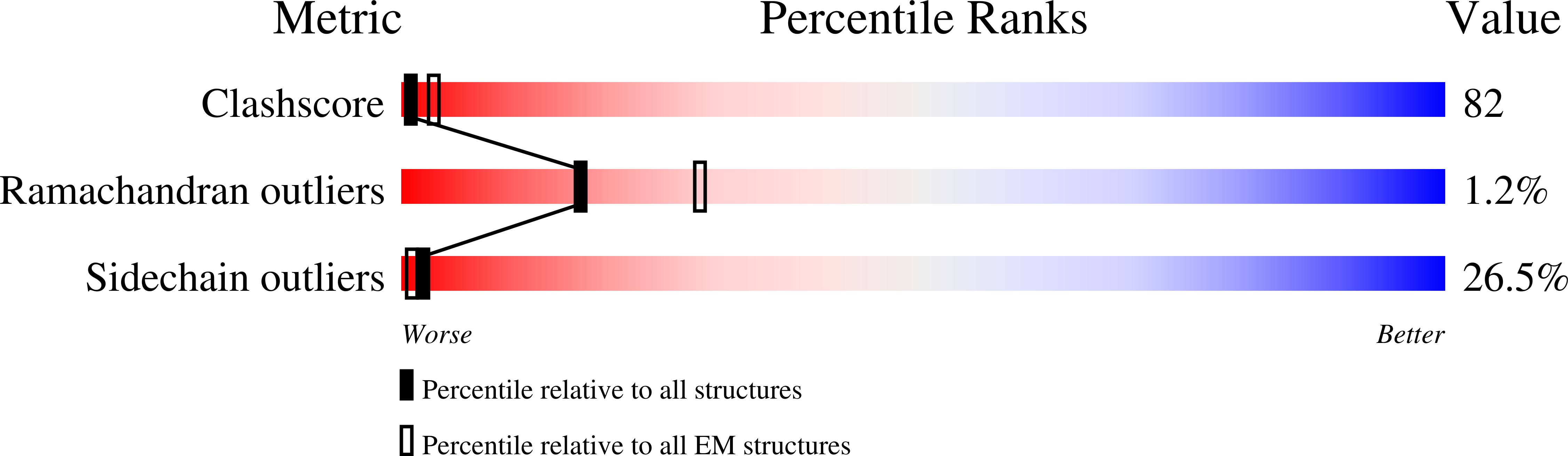
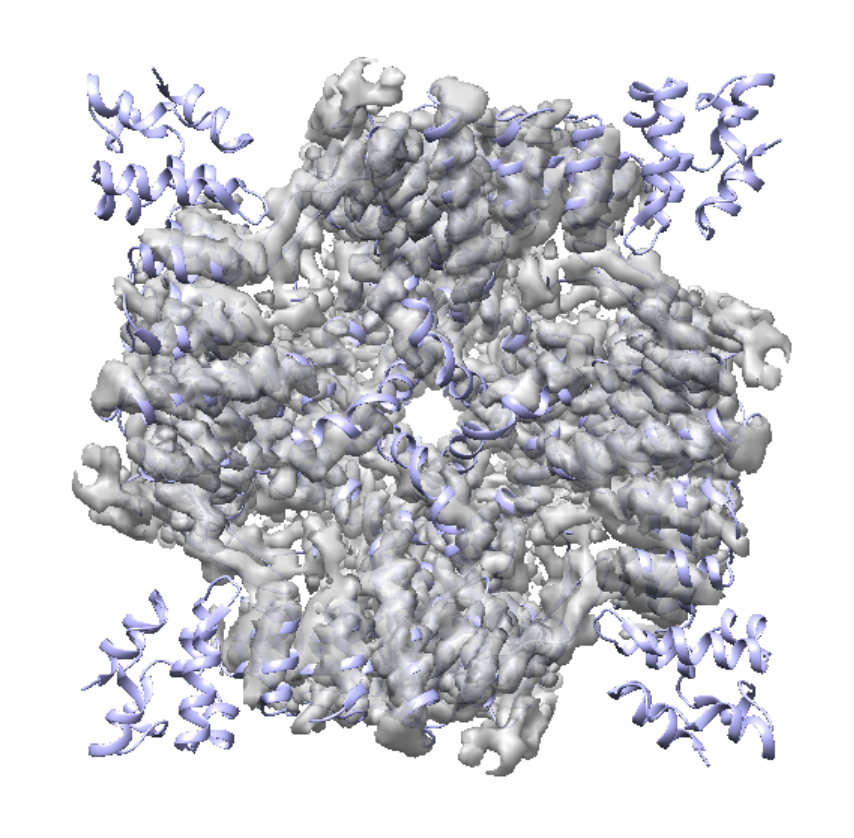
First, roughly fit the model into the map. This can be done in ChimeraX. They have slightly different conformation, so the fit won't be very precise. It is ok to leave it as is.
Compile model parameters
First, we compute some parameters from the input model that will be used in the following refinement.
e2gmm_model_compile.py --model 3j5q.pdb
This should create a gmm_model_xx folder, which contains information on the stereochemical constraints of the input model. All output will be in PDB format if the input model here is PDB. If the input is CIF, the output will be in CIF. Otherwise, specifying --writecif will force output to be CIF.
For your own data, it is worth noting that some non-protein structures are not supported at the moment. It is recommended to remove them from input and add them back after refinement. It is also good to notify me about those so I can add them in the future.
Fit to density map
e2gmm_model_fit.py --path gmm_model_00 --map emd_8117.mrc --resolution 3 --writetxt --rebuild_rotamer
Here gmm_model_00 is the folder generated by the e2gmm_model_comple.py command. This step can be skipped if there is no input map and you only want to refine the stereochemical score of the model.
Make sure the model does fit in the map in the first place. In this case, the loss from command line output should start from around -0.4 and gradually decrease to -0.6 or lower throughout iterations.
The --rebuild_rotamer option will select the rotamer for each residue that fits best into the given map (by map-model FSC). The process can be very slow. It is necessary for this case because there are many sidechain outliers, but can be skipped if the starting model is more reasonable.
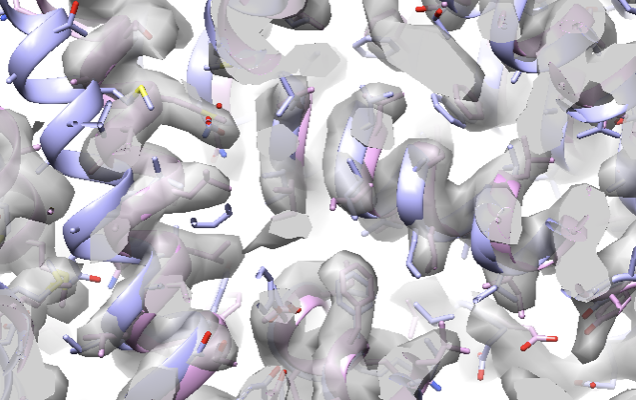
Take a look at fit_03.pdb and the model should fit into the map.
Full model refinement
e2gmm_model_refine.py --path gmm_model_00 --model gmm_model_00/fit_03.pdb
gmm_model_00/fit_03.pdb is produced by e2gmm_model_fit.py. If the fitting step is skipped, simply remove the --model option.
Normally, the default option should produce a good enough model. But in this case, since we started from a quite bad position, it would be necessary to run the command multiple times. For the second run, specify --niter 0,20 to skip the initial iterations, and --fixrotamer to force good rotamers for the bad residues.
e2gmm_model_refine.py --path gmm_model_00 --model gmm_model_00/model_02.pdb --fixrotamer --niter 0,20
There is some randomness in the process, and the command can be run multiple times. In the end, this should give relatively good PDB validation score.
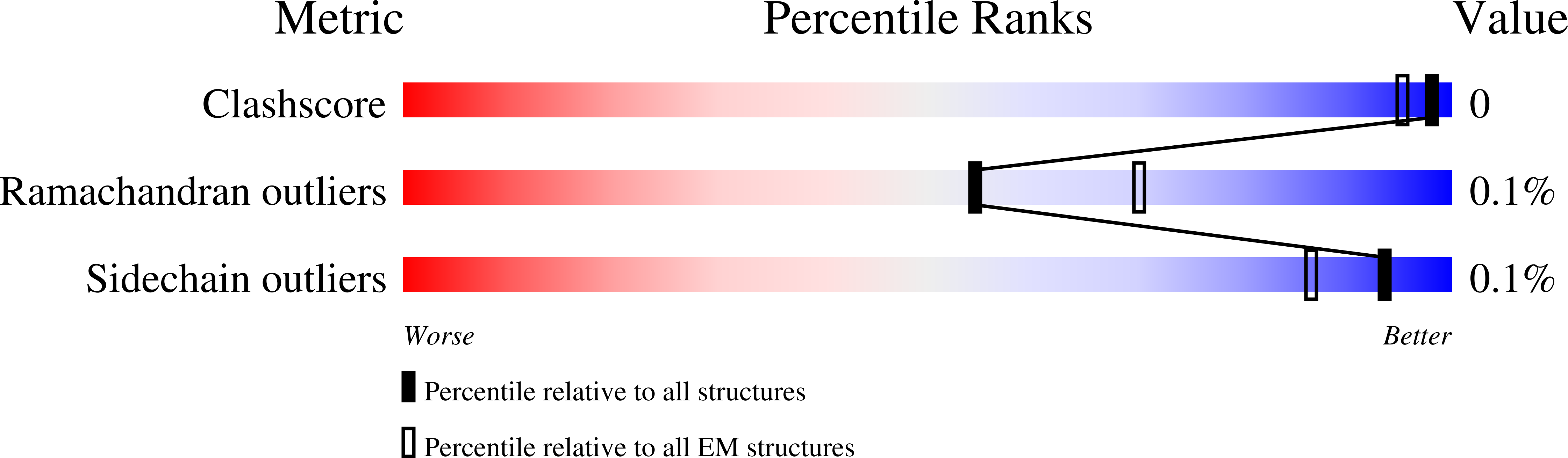
Multi-model refinement
After heterogeneity analysis, we can build models for a continuous conformational change using a stack of volumes generated by e2gmm_eval.py.
e2gmm_model_multi.py --path gmm_model_00 --modelpdb gmm_model_00/model_02.pdb --modeltxt gmm_model_00/fit_03.txt --maps gmm_00/ptcls_cls_01.hdf --resolution 10 --ndim 2