|
Size: 8134
Comment: draft
|
Size: 8492
Comment: update 10180
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 4: | Line 4: |
| Updates: * After 03/22/2021, a new GUI program, '''e2gmm_analysis''' is added to visualize the motion trajectory of the Gaussian model after training. * After 05/05/2021, a new '''--conv''' option is added that uses a convolutional neural network structure and improves convergence. |
|
| Line 69: | Line 72: |
| e2gmm_refine.py --model model_gmm.txt --ptclsin ptcls_small.hdf --heter --maxboxsz 48 --gradout allgrads.hdf --midout mid_00.txt --pas 010 | e2gmm_refine.py --model model_gmm.txt --ptclsin ptcls_small.hdf --heter --maxboxsz 48 --midout mid_00.txt --pas 010 |
| Line 81: | Line 84: |
| We use another dataset of pre-catalytic spliceosome from [[https://www.ebi.ac.uk/pdbe/emdb/empiar/entry/10180| EMPIAR-10180]] as an example of continuous conformational change. The sample dataset includes 20,000 particles with alignment information in their headers. Temporary download link [[https://drive.google.com/file/d/1VQGVGoLyBi4840pUGYy7Ougz-UF1DyEH | here]]. | We use another dataset of pre-catalytic spliceosome from [[https://www.ebi.ac.uk/pdbe/emdb/empiar/entry/10180| EMPIAR-10180]] as an example of continuous conformational change. The sample dataset includes 20,000 particles with alignment information in their headers. Temporary download link [[https://drive.google.com/file/d/1th4OP4McoZFWYQcxDzo4SkahawqvNNTq | here]]. Similar to the ribosome dataset, here we intentionally selected a subset of particles that follows a certain motion trajectory. Inside the zip file is a minimum single particle refinement project, including a '''particles''' folder that contains the downsampled particles, and a '''r3d_00''' folder with particle alignment information and the 3D average structure. There is also a '''strucfac.txt''' file that is used for optimal sharpening of the density maps. |
| Line 83: | Line 86: |
| First, create a folder to contain related files. {{{ |
|
| Line 84: | Line 89: |
e2project3d.py r3d_00/threed_01.hdf --outfile gmm_00/projections.hdf --orientgen=eman:delta=5 --parallel thread:12 e2gmm_refine.py --projs gmm_00/projections.hdf --npt 512 --maxboxsz 24 --modelout gmm_00/model_gmm.txt --niter 20 --mask r3d_00/mask.hdf e2gmm_refine.py --projs gmm_00/projections.hdf --npt 1024 --maxboxsz 64 --model gmm_00/model_gmm.txt --modelout gmm_00/model_gmm.txt --niter 20 --mask r3d_00/mask.hdf --evalmodel gmm_00/model_projs.hdf e2make3dpar.py --input gmm_00/model_projs.hdf --output gmm_00/model_avg.hdf --pad 128 --mode trilinear --keep 1 --threads 12 --setsf strucfac.txt e2proc3d.py gmm_00/model_avg.hdf gmm_00/fsc_map_model.txt --calcfsc r3d_00/threed_01.hdf e2gmm_refine.py --model gmm_00/model_gmm.txt --ptclsin r3d_00/ptcls_01.lst --heter --maxboxsz 64 --gradout gmm_00/allgrads.hdf --midout gmm_00/mid_00.txt --conv ptcl_pca_kmean_m3d.py --pts gmm_00/mid_00.txt --pcaout gmm_00/mid_pca.txt --ptclsin r3d_00/ptcls_01.lst --ptclsout gmm_00/ptcls_cls --setsf strucfac.txt --mode regress --ncls 9 We can use the same command to reconstruct the averaged map and build the neutral Gaussian model. Note that due to the difference in magnification, at the Fourier box size of 48, we are only at ~24 Å resolution in this dataset, so make sure to filter the maps accordingly. For the heterogeneity analysis, run {{{ e2gmm_refine.py --model model_gmm.txt --ptclsin ptcls_small.hdf --heter --maxboxsz 48 --gradout allgrads.hdf --midout mid_00.txt --pas 110 |
|
| Line 106: | Line 91: |
| Note that this time we set --pas to 110, so both the position and amplitude of the Gaussian functions are allowed to change to minimize particle-projection difference. Since we are looking at a continuous motion, instead of classification, we reconstruct 3D maps along the first eigenvector from PCA with even spacing. |
Generate projections of the averaged structure: |
| Line 109: | Line 93: |
| ptcl_pca_kmean_m3d.py --pts mid_00.txt --pcaout mid_pca.txt --ptclsin ptcls_small.hdf --ptclsout ptcls_cls --setsf sf_lp.txt --mode regress --ncls 4 | e2project3d.py r3d_00/threed_01.hdf --outfile gmm_00/projections.hdf --orientgen=eman:delta=5 --parallel thread:12 |
| Line 112: | Line 96: |
| Due to the limited amount of particles, the motion observed from this sample subset won't be as significant as the heterogeneity from the full dataset, but the basic motion mode can already be visualized. | Now build the Gaussian model. We can start from lower resolution and fewer Gaussian functions, then increase both to have better convergence. {{{ e2gmm_refine.py --projs gmm_00/projections.hdf --npt 512 --maxboxsz 24 --modelout gmm_00/model_gmm.txt --niter 20 --mask r3d_00/mask.hdf e2gmm_refine.py --projs gmm_00/projections.hdf --npt 1024 --maxboxsz 64 --model gmm_00/model_gmm.txt --modelout gmm_00/model_gmm.txt --niter 20 --mask r3d_00/mask.hdf --evalmodel gmm_00/model_projs.hdf }}} Check the Gaussian model and make sure it is consistent with the density map. {{{ e2make3dpar.py --input gmm_00/model_projs.hdf --output gmm_00/model_avg.hdf --pad 128 --mode trilinear --keep 1 --threads 12 --setsf strucfac.txt e2proc3d.py gmm_00/model_avg.hdf gmm_00/fsc_map_model.txt --calcfsc r3d_00/threed_01.hdf }}} Now perform the heterogeneity analysis. Here use the new '''--conv''' mode to speed up convergence. Here we look at both movement trajectory and amplitude change, so the '''--pas''' option can be omitted. {{{ e2gmm_refine.py --model gmm_00/model_gmm.txt --ptclsin r3d_00/ptcls_01.lst --heter --maxboxsz 64 --midout gmm_00/mid_00.txt --conv }}} Finally, classify particles based on their distribution in the conformational space. Since this is a continuous motion, we use '''--mode=regress''' to generate 3D density maps along the first eigenvector at a constant spacing. {{{ ptcl_pca_kmean_m3d.py --pts gmm_00/mid_00.txt --pcaout gmm_00/mid_pca.txt --ptclsin r3d_00/ptcls_01.lst --ptclsout gmm_00/ptcls_cls --setsf strucfac.txt --mode regress --ncls 9 }}} |
Structural variability analysis with Gaussian mixture model
Available in EMAN release after 10/01/2020. Still under development. For details of the method, refer to Chen2021. Updates:
After 03/22/2021, a new GUI program, e2gmm_analysis is added to visualize the motion trajectory of the Gaussian model after training.
After 05/05/2021, a new --conv option is added that uses a convolutional neural network structure and improves convergence.
Dataset
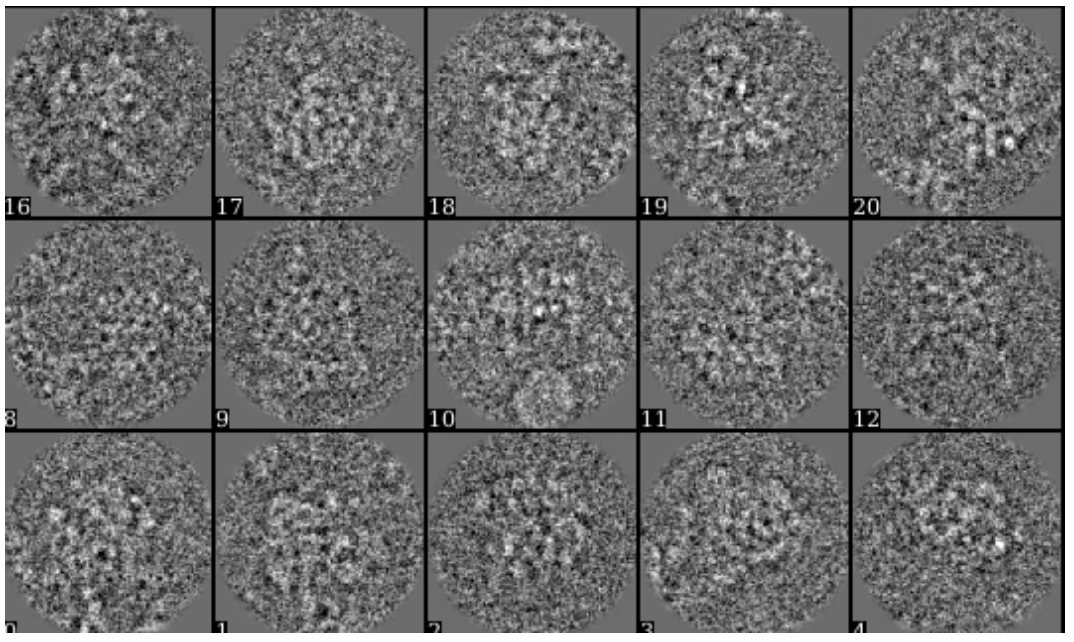
First, for the compositional heterogeneity analysis, we use the dataset of L17-Depleted 50S Ribosomal Intermediates from EMPIAR-10076. In this tutorial, we use a subset of only 6000 particles of box size 128. The small dataset can be downloaded here. The particles are artificially selected to keep the tutorial is as simple as possible. To skip the alignment step, a single model refinement is run with the full dataset and the orientation of each particle is saved in the header of the image (xform.projection).
If you have previously run e2refine_easy on this data, you can extract the necessary files from a refine_XX folder using e2evalrefine.py with the --extractorientptcl option, and then just use the corresponding threed_XX file as a starting volume for the analysis.
The single model refinement can also be performed with the new e2spa_refine.py pipeline, which directly outputs the aligned list file and average density map that can be used as input.
To convert a refinement from Relion, a script called e2convertrelion.py can be found in the examples folder (after 01/2021) that produces phase flipped particles and aligned list file.
Reconstruct density map
Skip this step if you have an averaged map already. For the short tutorial, it is a useful step to double-check all the information in the particles is correct.
First, make the 3D map using the particles (or list file) with alignment information.
e2make3dpar.py --input ptcls_small.hdf --output avg.hdf --pad 192 --mode trilinear --keep 1 --threads 12
Now do a quick map sharpening for the output. The sharpened then filtered map is saved as avgsf.hdf. Also, create a structure factor file for the filtered map to simplify future reconstruction processes.
e2spt_structfac.py avg.hdf --res 15 e2proc3d.py avgsf.hdf avgsf.hdf --calcsf sf_lp.txt
Then make a mask and apply it to the map. The mask will be used to exclude Gaussian functions outside the boundary later. Now we have a “neutral” structure to start with.
e2proc3d.py avgsf.hdf msk.hdf --process=normalize.edgemean: --process=mask.auto3d.thresh:nshells=4:nshellsgauss=8:threshold1=5:threshold2=2:return_mask=1 e2proc3d.py avgsf.hdf avgmsk.hdf --process=normalize.edgemean --multfile msk.hdf
Build the Gaussian model
To limit memory consumption, the model is trained with 2D projections of the model instead of the 3D map. So here we make projections of the averaged map first.
e2project3d.py avgmsk.hdf --outfile avg_projs.hdf --orientgen=eman:delta=5 --parallel thread:12
Now we begin training the Gaussian model from scratch. We start from a tiny box size so it converges faster. Then run with a Fourier box size of 48 so the Gaussian model matches the density map at ~15 Å. The mask will exclude the Gaussian centers outside the volume. The second command can be repeated if the Gaussian model does not match well with the density map.
e2gmm_refine.py --projs avg_projs.hdf --npt 2048 --maxboxsz 24 --modelout model_gmm.txt --niter 20 --mask msk.hdf e2gmm_refine.py --projs avg_projs.hdf --maxboxsz 48 --modelout model_gmm.txt --niter 20 --evalmodel model_projs.hdf --model model_gmm.txt --npt 2048 --mask msk.hdf
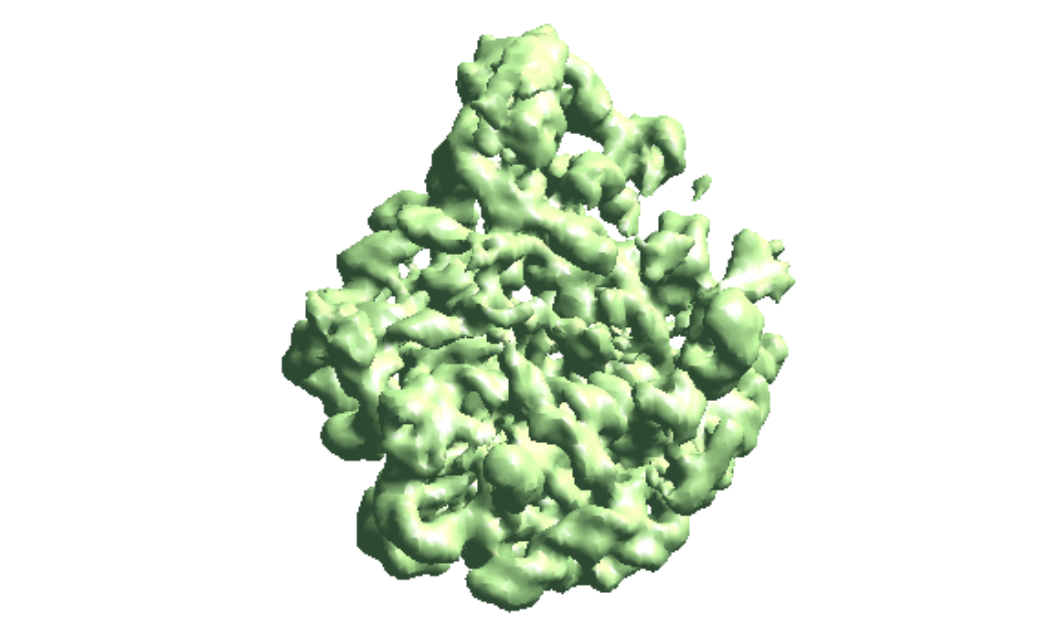
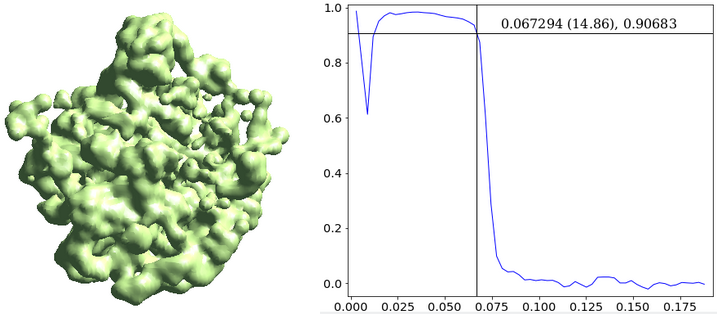
The --evalmodel option of e2gmm_refine.py will write the projection of the model to an image stack so we can take a look at the 3D reconstruction and check its FSC with the input density map.
e2make3dpar.py --input model_projs.hdf --output model_avg.hdf --pad 192 --mode trilinear --keep 1 --threads 12 --setsf sf_lp.txt e2proc3d.py model_avg.hdf fsc_00.txt --calcfsc avgmsk.hdf
Heterogeneity analysis
Now we have a Gaussian model of the neutral structure, we can look into the heterogeneity of the particles. Run the e2gmm_refine.py command with particle input and --heter option to let the program embed them to a conformation space. The --gradout option writes the gradient with respect to the neutral structure to a file so the step can be skipped next time (read the saved file using --gradin). The middle layer output is saved in mid_00.txt. The --pas option controls the adjustment of position, amplitude, and sigma of the Gaussian functions. Setting it to 010 means changing amplitude only.
e2gmm_refine.py --model model_gmm.txt --ptclsin ptcls_small.hdf --heter --maxboxsz 48 --midout mid_00.txt --pas 010
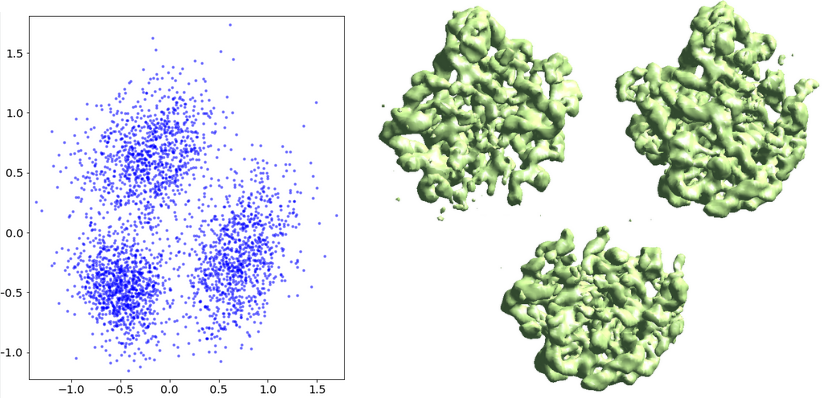
Finally, the ptcl_pca_kmean_m3d.py script (in examples folder) runs PCA on the middle layer output, classifies the particles, and reconstructs 3D maps of each class.
ptcl_pca_kmean_m3d.py --pts mid_00.txt --pcaout mid_pca.txt --ptclsin ptcls_small.hdf --ptclsout ptcls_cls --setsf sf_lp.txt
Continuous heterogeneity
We use another dataset of pre-catalytic spliceosome from EMPIAR-10180 as an example of continuous conformational change. The sample dataset includes 20,000 particles with alignment information in their headers. Temporary download link here. Similar to the ribosome dataset, here we intentionally selected a subset of particles that follows a certain motion trajectory. Inside the zip file is a minimum single particle refinement project, including a particles folder that contains the downsampled particles, and a r3d_00 folder with particle alignment information and the 3D average structure. There is also a strucfac.txt file that is used for optimal sharpening of the density maps.
First, create a folder to contain related files.
mkdir gmm_00
Generate projections of the averaged structure:
e2project3d.py r3d_00/threed_01.hdf --outfile gmm_00/projections.hdf --orientgen=eman:delta=5 --parallel thread:12
Now build the Gaussian model. We can start from lower resolution and fewer Gaussian functions, then increase both to have better convergence.
e2gmm_refine.py --projs gmm_00/projections.hdf --npt 512 --maxboxsz 24 --modelout gmm_00/model_gmm.txt --niter 20 --mask r3d_00/mask.hdf e2gmm_refine.py --projs gmm_00/projections.hdf --npt 1024 --maxboxsz 64 --model gmm_00/model_gmm.txt --modelout gmm_00/model_gmm.txt --niter 20 --mask r3d_00/mask.hdf --evalmodel gmm_00/model_projs.hdf
Check the Gaussian model and make sure it is consistent with the density map.
e2make3dpar.py --input gmm_00/model_projs.hdf --output gmm_00/model_avg.hdf --pad 128 --mode trilinear --keep 1 --threads 12 --setsf strucfac.txt e2proc3d.py gmm_00/model_avg.hdf gmm_00/fsc_map_model.txt --calcfsc r3d_00/threed_01.hdf
Now perform the heterogeneity analysis. Here use the new --conv mode to speed up convergence. Here we look at both movement trajectory and amplitude change, so the --pas option can be omitted.
e2gmm_refine.py --model gmm_00/model_gmm.txt --ptclsin r3d_00/ptcls_01.lst --heter --maxboxsz 64 --midout gmm_00/mid_00.txt --conv
Finally, classify particles based on their distribution in the conformational space. Since this is a continuous motion, we use --mode=regress to generate 3D density maps along the first eigenvector at a constant spacing.
ptcl_pca_kmean_m3d.py --pts gmm_00/mid_00.txt --pcaout gmm_00/mid_pca.txt --ptclsin r3d_00/ptcls_01.lst --ptclsout gmm_00/ptcls_cls --setsf strucfac.txt --mode regress --ncls 9