|
Size: 972
Comment:
|
Size: 2231
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 4: | Line 4: |
To see the entire list of command line options for e2tomoboxer (not many at this point), and an explanation of what they're for, type ''e2tomoboxer.py -h'' at the command line. |
|
| Line 14: | Line 16: |
| {{{ e2tomoboxer.py tomogram.rec --yshort --inmemory }}} If you don't provide the ''--inmemory'' option, the data will be read from disk, opposed to being read from memory (which is faster). |
|
| Line 15: | Line 21: |
| {{{ e2tomoboxer.py tomogram.rec --yshort --inmemory }}} If you don't provide the "inmemory" option, the data will be read from disk, opposed to being read from memory (which is faster). |
Provide the ''--yshort'' option if the shortest dimension of your tomogram (the "ice thickness") is along the y axis (for example, 4096 x 500 x 4096). |
| Line 19: | Line 23: |
| Provide the "yshort" option if the shortest dimension of your tomogram (the "ice thickness") is along the y axis (for example, 4096 x 500 x 4096). | e2tomoboxer consists of _3 GUI windows_. 1) The __''MAIN'' window__ opens immediately when you run ''e2tomoboxer.py'' from the command line. 2) The __''PARTICLE LIST'' window__ opens when you click anywhere in the slice views of the MAIN window to select a subvolume. 3) Same thing for the __''SINGLE PARTICLE VIEW'' window__. '''MAIN tomoboxer window''' This window consists of three orthogonal views or slices through the tomogram, parallel to the XY, XZ and YZ planes, and a small options panel on the bottom left corner. {{attachment:main.png}} The yellow cross that comes up when you click on any of the views indicates the position of the cutting planes. You can "scroll" through the slices in the largest projection (XY, looking at the tomogram from the "top") with the scroll bar on the bottom right corner, or by scrolling with the middle mouse button while pressing and holding SHIFT. ''' PARTICLE LIST window''' {{attachment:particle_list.png}} '''SINGLE PARTICLE VIEW window''' {{attachment:single_particle_viewpng}} |
e2tomoboxer
This program is used for interactive tomographic single particle picking (locating and extracting subtomograms).
To see the entire list of command line options for e2tomoboxer (not many at this point), and an explanation of what they're for, type e2tomoboxer.py -h at the command line.
Options
-B |
--boxsize |
int |
Box size in pixels |
|
--inmemory |
bool |
This will read the entire tomogram into memory. Much faster, but you must have enough ram ! |
|
--yshort |
bool |
This means you have a file where y is the short axis |
|
--apix |
float |
Override the A/pix value stored in the tomogram header |
-h |
--help |
bool |
show this help message and exit |
-v |
--verbose |
int |
verbose level [0-9], higner number means higher level of verboseness |
USAGE
e2tomoboxer.py tomogram.rec --yshort --inmemory
If you don't provide the --inmemory option, the data will be read from disk, opposed to being read from memory (which is faster).
Provide the --yshort option if the shortest dimension of your tomogram (the "ice thickness") is along the y axis (for example, 4096 x 500 x 4096).
e2tomoboxer consists of _3 GUI windows_.
1) The MAIN window opens immediately when you run e2tomoboxer.py from the command line.
2) The PARTICLE LIST window opens when you click anywhere in the slice views of the MAIN window to select a subvolume.
3) Same thing for the SINGLE PARTICLE VIEW window.
MAIN tomoboxer window
This window consists of three orthogonal views or slices through the tomogram, parallel to the XY, XZ and YZ planes, and a small options panel on the bottom left corner.
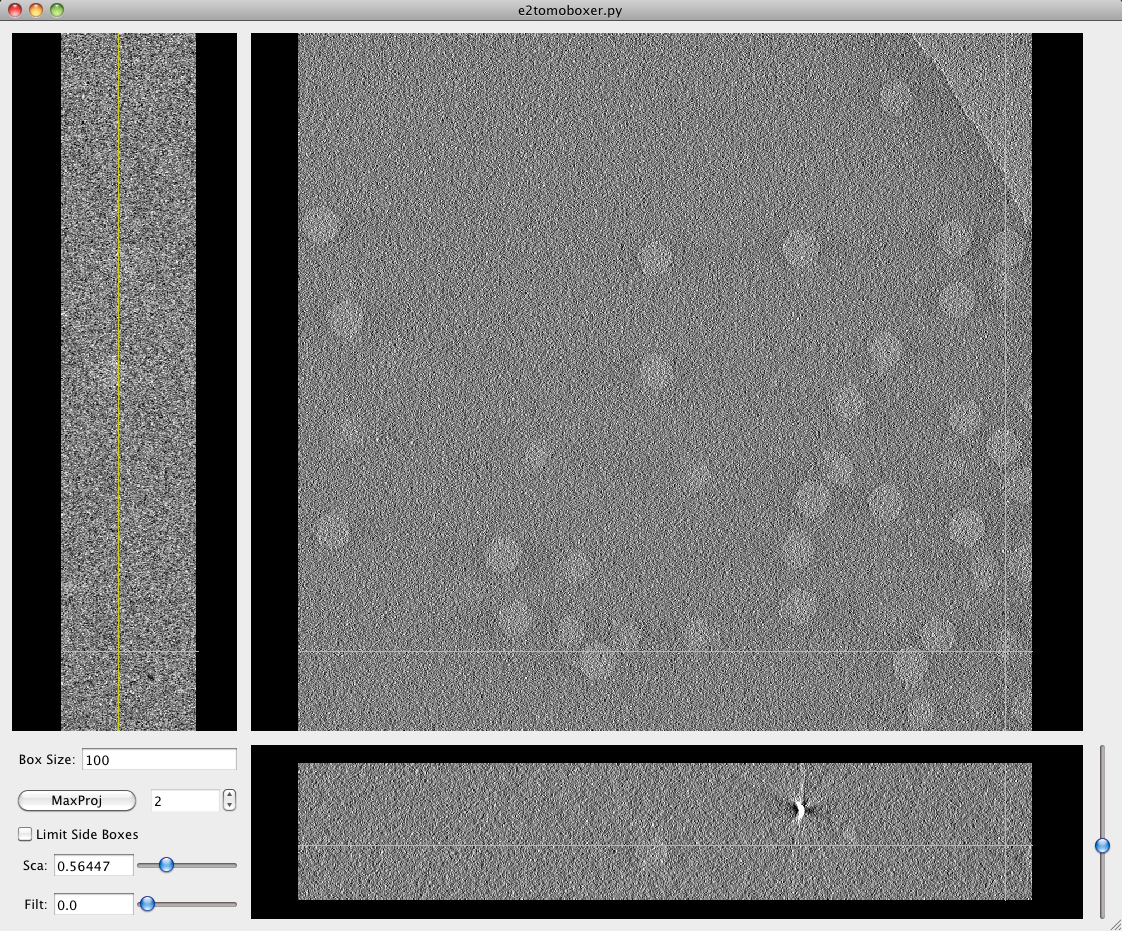
The yellow cross that comes up when you click on any of the views indicates the position of the cutting planes.
You can "scroll" through the slices in the largest projection (XY, looking at the tomogram from the "top") with the scroll bar on the bottom right corner, or by scrolling with the middle mouse button while pressing and holding SHIFT.
PARTICLE LIST window

SINGLE PARTICLE VIEW window
