|
Size: 3324
Comment:
|
Size: 3325
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 36: | Line 36: |
| e2helixboxer.py <options (not --gui)> <micrograph> Options: | e2helixboxer.py <options (not --gui)> <micrograph> Options: |
e2helixboxer.py: Overview
e2helixboxer.py is used to select rectangular 2D projections of helices from a micrograph, and extract overlapping particles from the boxed helices. The boxing must be done in GUI mode, but extracting particles from boxed regions may be done from the GUI or from the command line.
GUI mode
To start the program's graphic user interface, use the "--gui" option. You can follow this with a micrograph filename, a list of micrograph filenames, or nothing.
$ e2helixboxer.py --gui <micrograph1> <micrograph2> <...>
$ e2helixboxer.py --gui $ e2helixboxer.py --gui 101.mrc $ e2helixboxer.py --gui *.mrc micrograph.hdf *.img abc.dm3
The left window is the helix viewer, the middle is the micrograph viewer, and the right is the main window.
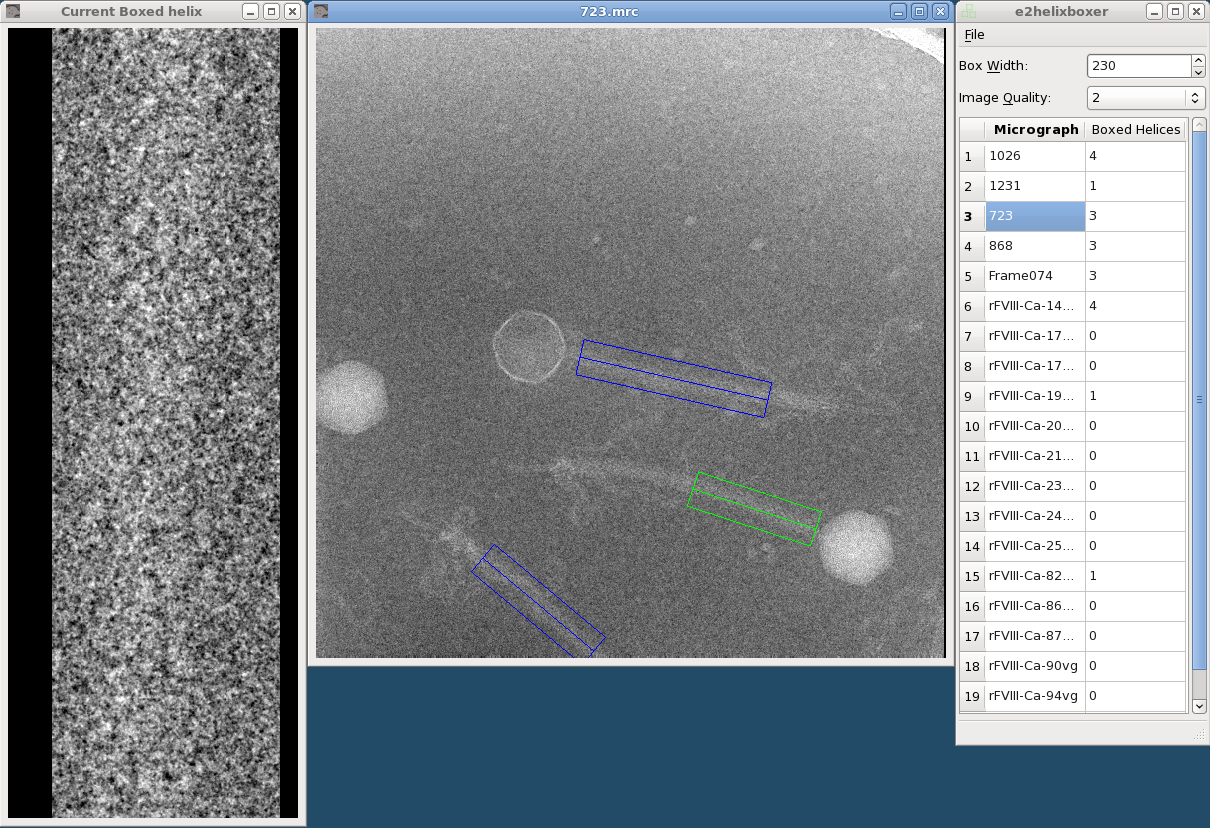
The main window shows a list of open micrographs and how many helices you have boxed in each one. (Actually, only one micrograph is loaded in memory at a time, but this allows for quick switching between particles.) Any micrographs you specified after the "--gui" option will be listed.
Box Editing |
|
Draw box |
Left click and drag |
Move box |
Left click near box center and drag |
Move one box endpoint |
Left click near box end and drag |
Delete box |
Hold shift and left click inside box |
After you have boxed the helices in a micrograph, you can go to "File->Save" to save the coordinates and image data of the helices and the particles you extract from them.
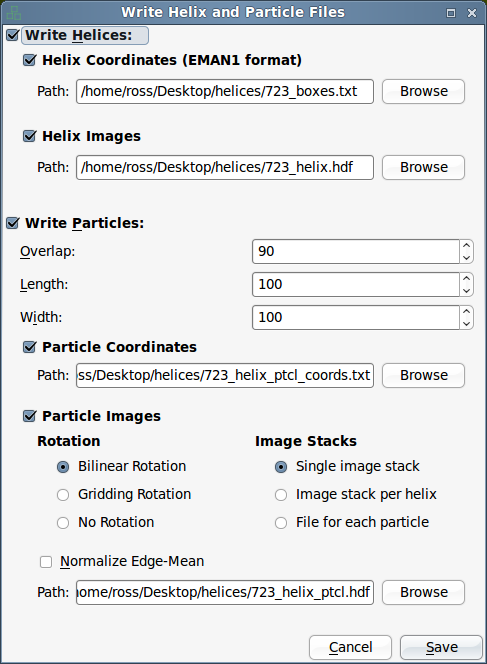
Command-line mode
In command-line mode, you can only work with one micrograph at a time. You should have already boxed particles in the GUI mode; if that is done the box coordinates should be saved in the EMAN2 database.
e2helixboxer.py <options (not --gui)> <micrograph> Options:
- --version show program's version number and exit -h, --help show this help message and exit --gui Start the graphic user interface for boxing helices. -X HELIX_COORDS, --helix-coords=HELIX_COORDS
- Save coordinates for helices in EMAN1 *.box format: x1-w/2 y1-w/2 w w -1 x2-w/2 y2-w/2 w w -2
- Save images of the helices. The file name specified will have helix numbers added to it.
- Save coordinates of the centers of particles with the specified overlap in pixels
- Save images of the particles with the specified overlap in pixels. The file name specified will have helix numbers (and particle numbers if the file type does not support image stacks) added to it.
- Helix width in pixels
- Particle overlap in pixels
- Particle length in pixels
- Particle width in pixels
- square with length max(ptcl_length, ptcl_width).
- Apply the normalize.edgemean processor to each particle.
