| Display Basics | EMAN2 File Browser | 2D Display | 2D Stack Display | 3D Image Display | Euler Tool | e2display FAQ |
e2eulerxplor: EMAN2's EulerXplor Tool
|
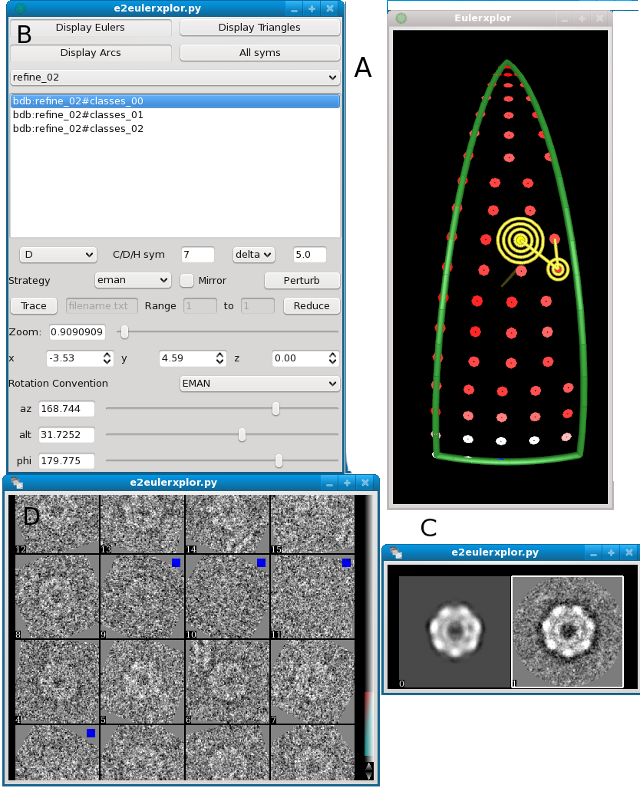
The four interfaces in e2eulerxplor. See the table below for more details |
Figure Label |
Description |
A |
This is a graphical depiction of orientations on the asymmetric unit. Click on one of the dots to display the associated class average and projection as shown in C. The color of the dot in the asymmetric unit shows the relative number of particles in the class: Blue indicates a class with a lot of particles in it, red indicates a class with few particles in it. White is in between. |
B |
This is main inspector, this appears when you middle click on A. You can select different refinement iterations to investigate how the class averages and aligned particles are changing through time. |
C |
Projection (left) and class average corresponding to selected orientation (yellow) in A |
D |
Aligned particles that formed the class average shown in C (right). If you select the projection in C (left) these particles will be shown unaligned. Blue squares indicate that the particle did not make it into the final class average |
Mouse Handling
Mouse Action |
Result |
left button release |
Select a cylinder/dot in A, resulting in update of the image in B (below) or C above |
Right button drag |
x and y movement |
Shift + right button drag |
z movement |
Mouse wheel in and out |
Zoom in and out, respectively |
Left button drag |
Object rotation, quaternion based |
Middle button |
Show inspector (B) |
Inspecting EMData lists with euler_display from e2.py
You can open a list of EMData's with the EulerXplor tool if and only if they all have the xform.projection header attribute. Here is an example:
Note that all projection and class average images produced by e2workflow have the xform.projection header attribute, which are located in directories called"refine_??" - you're safe to try this out using those images.
|
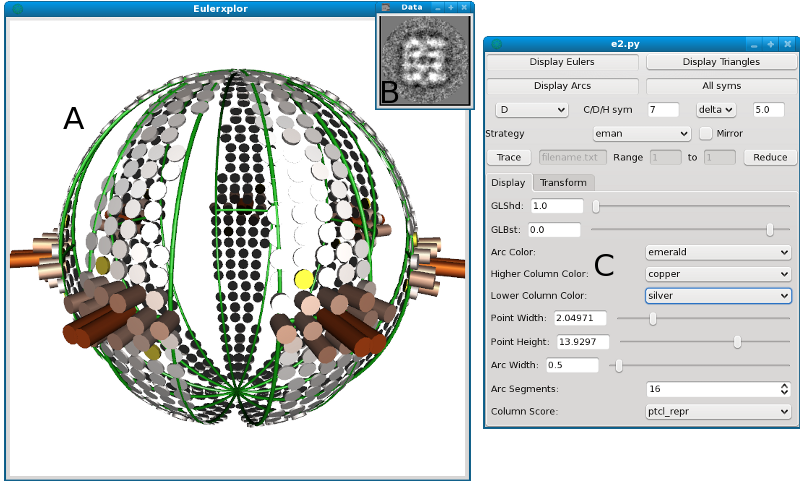
Typical screenshot of the Eulerxplor tool, launched using euler_display from e2.py |
Figure Label |
Description |
A |
Symmetrized orientation distribution. Cylinder heights are proportional to the "ptcl_repr" header attribute. You can change the header attribute that is represented by the cylinder height in the inspector (bottom of C) |
B |
The image corresponding to the selected (yellow) cylinder. |
C |
The inspector. Note you can uniformly enlarge the displayed cylinders using the "Point Height" and "Point Width" sliders |
Specialized Mouse Handling
When you open a list of EMData objects from Python Eulerxplor handles mouse events slightly differently. The only way to view an image associated with a point on the sphere is to hold shift and click on it.
Mouse Action |
Result |
Shift + left click |
Select a cylinder in A, resulting in update of the image in B |
Inspecting Transform lists with euler_display from e2.py
Similarly you can look at a list of Transforms with the EulerXplor tool, using this syntax: